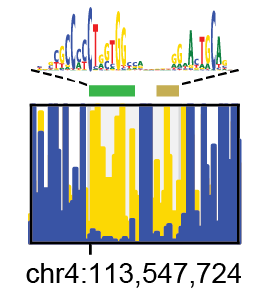
Non-redundant TF motif matches genome-wide
The redundancy and overlap between various motif model databases complicate downstream analysis and interpretation. We computed the pairwise similarity for >2,000 motif models determined for both human and mouse TFs and clustered them into 286 distinct motif clusters. Within each cluster we aligned motifs to each other to generate an archetypal consensus motif.